Analysis and Application of Chemical Modification Strategies for Small Nucleic Acid Drugs
RNA Interference (RNAi) is a gene-silencing mechanism first discovered in Caenorhabditis elegans, triggered by double-stranded small interfering RNA (siRNA), which induces sequence-specific gene silencing. In cells, siRNA is assembled into the RNA-induced silencing complex (RISC), which searches for and degrades complementary mRNA, thereby preventing the translation of target proteins. The development of siRNA drugs involves several key stages, including proof of concept (POC), clinical POC, and regulatory approval.
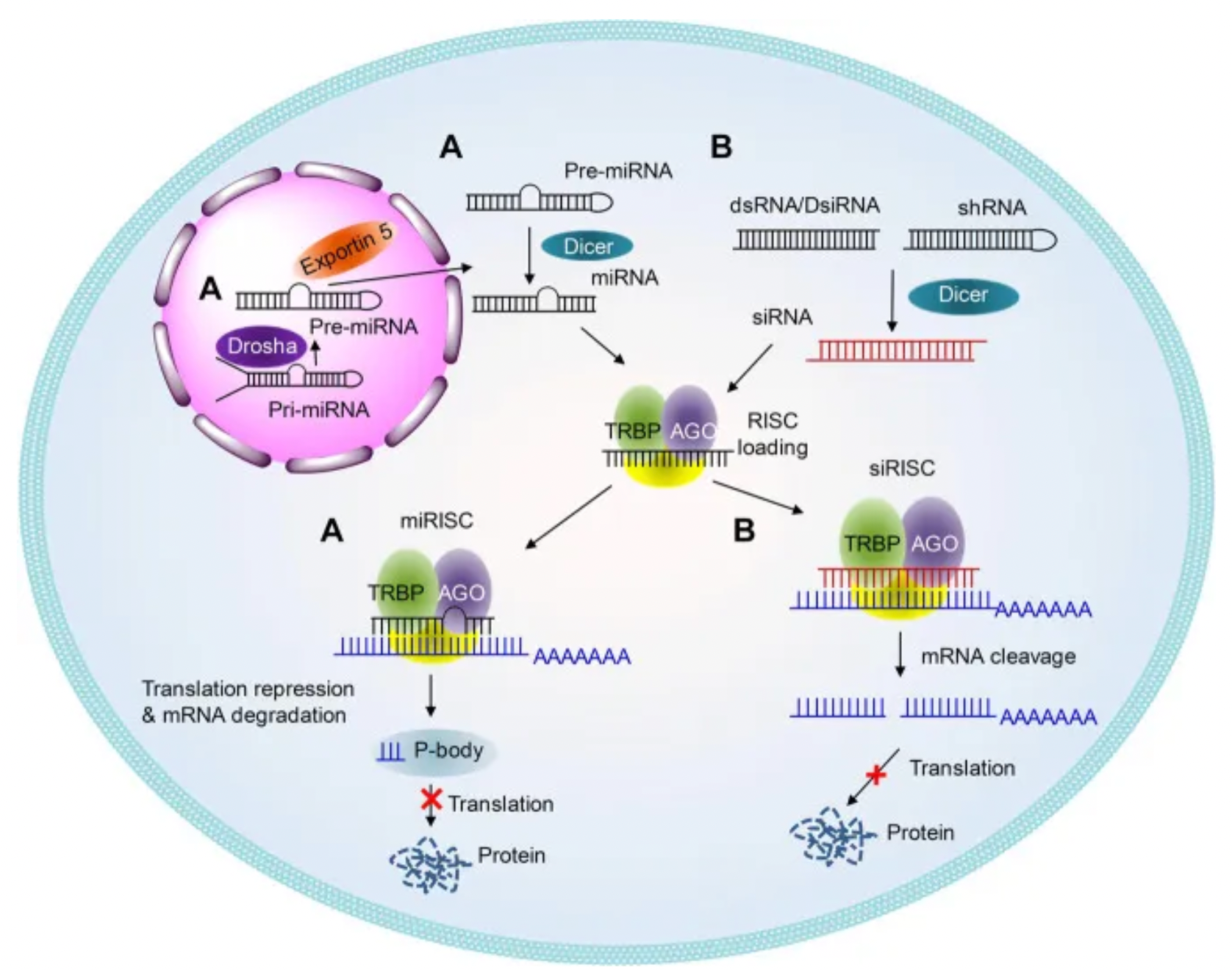
Chemical modifications of siRNA are crucial for enhancing stability, reducing immunogenicity and off-target effects, and improving cellular uptake. These modifications include phosphate backbone modifications, ribose modifications, and base modifications, which increase siRNA's resistance to nucleases, enhance its affinity for target mRNA, and reduce immune responses and nonspecific binding. Approved siRNA drugs, such as Patisiran, Givosiran, Lumasiran, Inclisiran, and Vutrisiran, employ specific chemical modifications and delivery systems to improve stability and efficacy. These chemical modifications help siRNA resist nuclease degradation, increase its affinity for target mRNA, and minimize immune responses and off-target effects.
In clinical applications, siRNA drugs exhibit unique pharmacokinetic (PK) and pharmacodynamic (PD) properties, characterized by short plasma exposure, prolonged target organ exposure, and sustained pharmacological effects. The key pharmacodynamic mechanism is the siRNA and RISC complex in the cytoplasm, rather than the total siRNA concentration in plasma or target organs. Therefore, nonclinical pharmacodynamic studies and proof-of-concept trials of siRNA drugs are essential for understanding their mechanisms of action and guiding clinical trial design.
Patsnap Bio, a powerful biosequence retrieval and analysis system, offers comprehensive coverage and deep processing of global protein and nucleic acid sequence data. The database enhances sequence information retrieval by manually annotating key biological sequences and marking modification structures. It also uses knowledge graphs to connect biological sequences with species, drugs, diseases, structures, and functions, providing researchers with a comprehensive resource to support siRNA drug discovery and development. In the development of siRNA drugs, Patsnap Bio helps researchers understand the global research status of specific sequences, perform patentability analysis and freedom-to-operate (FTO) assessments, and access the global research landscape based on target, drug, and sequence attributes. These functions are critical for nonclinical pharmacodynamic research and proof-of-concept trials, aiding in understanding mechanisms of action and guiding clinical trial design.
Next, we'll explore the effective chemical modification strategies used in approved siRNA drugs through Patsnap Bio.
Optimization of siRNA Drug Scaffolds
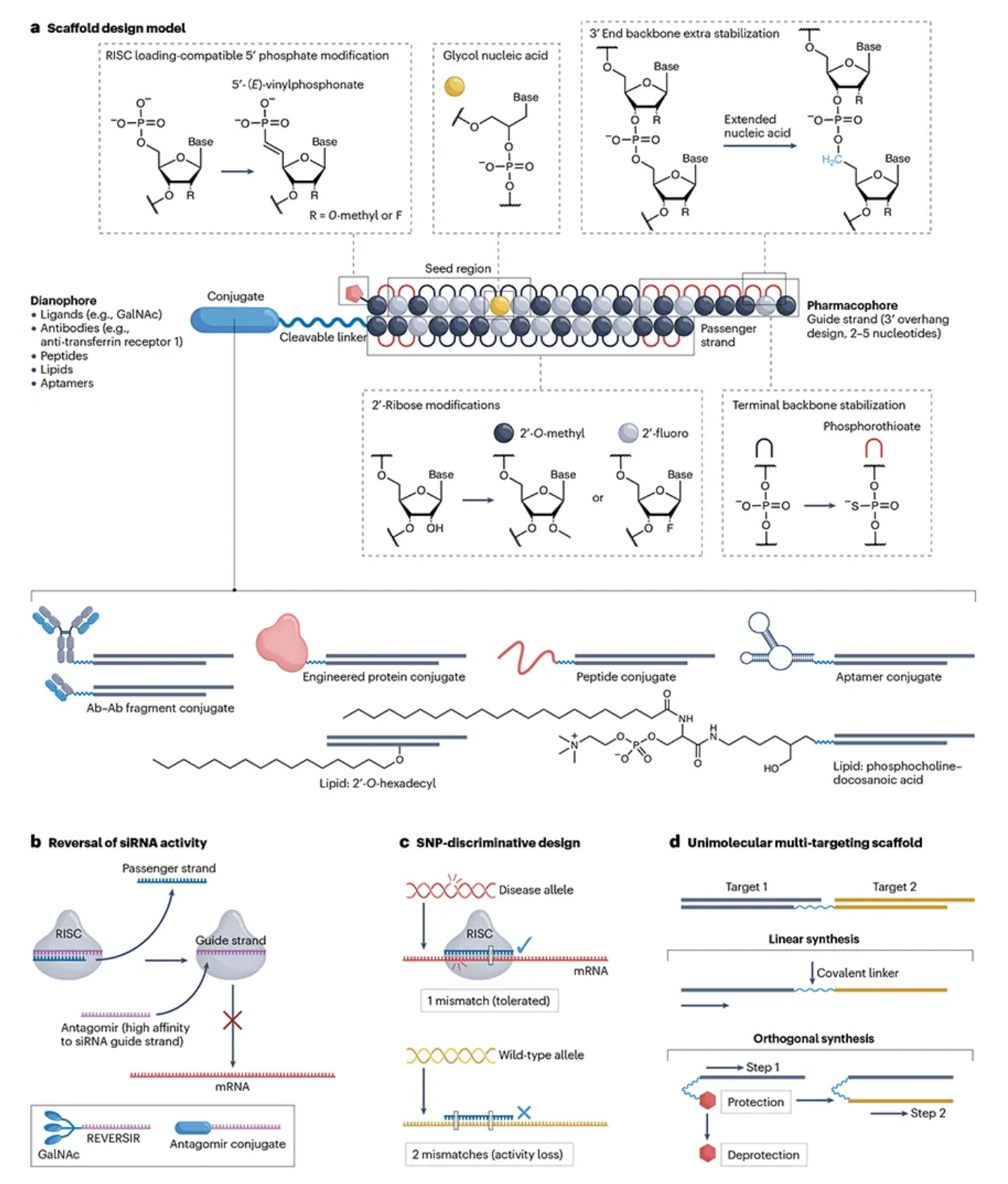
The stability of the basic scaffold of siRNA drugs is crucial to prevent metabolic degradation during tissue distribution and intracellular encapsulation. To optimize the interaction of siRNA with cellular processes, scientists have developed various chemical strategies, including modifications to nucleobases, ribose, and the backbone. Through Patsnap Bio, we identified that these modifications mainly include 2'-O-methyl (2'-OMe), 2'-fluoro (2'-F), and phosphorothioate (PS), which enhance siRNA’s resistance to nucleases, reduce immunogenicity and off-target effects, and increase affinity for target mRNA.
2'-OMe and 2'-F modifications protect the 2'-hydroxyl (2'-OH) from hydrolysis, maintaining siRNA stability, while PS modifications increase siRNA hydrophobicity, improving cellular uptake and potency. Although 2'-F modifications enhance hydrophobicity, safety concerns have arisen, particularly during Revusiran’s Phase III trial due to clinical toxicity issues. However, subsequent toxicological studies have shown that long-term administration of 2'-F modified siRNA is safe. While higher levels of 2'-F metabolites were detected in Revusiran compared to next-generation ESC compounds, no direct toxicity related to these metabolites was identified.
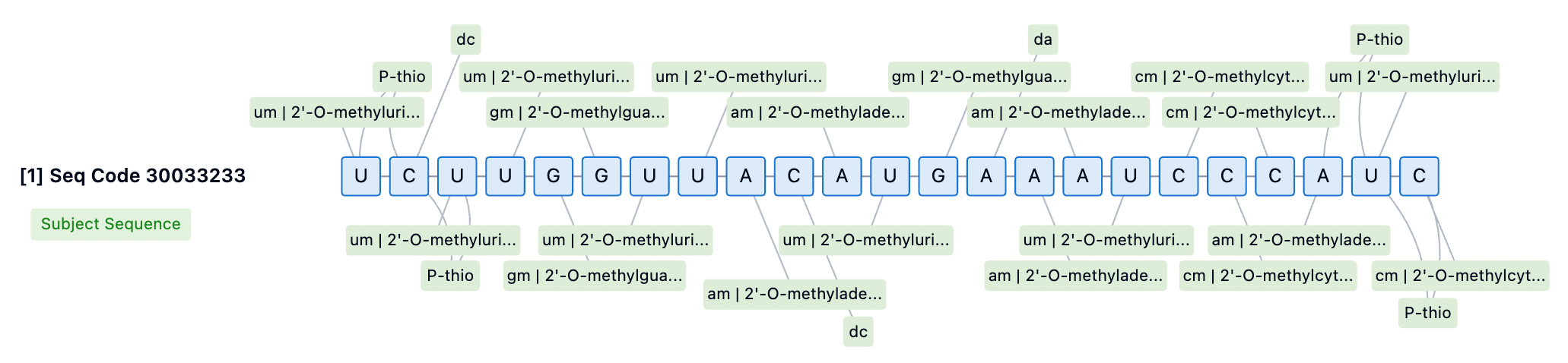
In siRNA drug design, the position and type of chemical modification significantly affect activity. For instance, 2'-OMe at specific positions in the guide strand may reduce activity. Therefore, identifying the optimal siRNA configuration requires iterative screening of sequences and modification patterns. Currently, fully 2'-modified siRNA structures often employ alternating 2'-OMe and 2'-F scaffolds, providing higher levels of stability and in vivo efficacy. Patsnap Bio revealed that the Lumasiran sequence utilizes an alternating 2'-OMe and 2'-F scaffold, enhancing its binding affinity to target mRNA and thereby improving the efficacy and specificity of the siRNA.
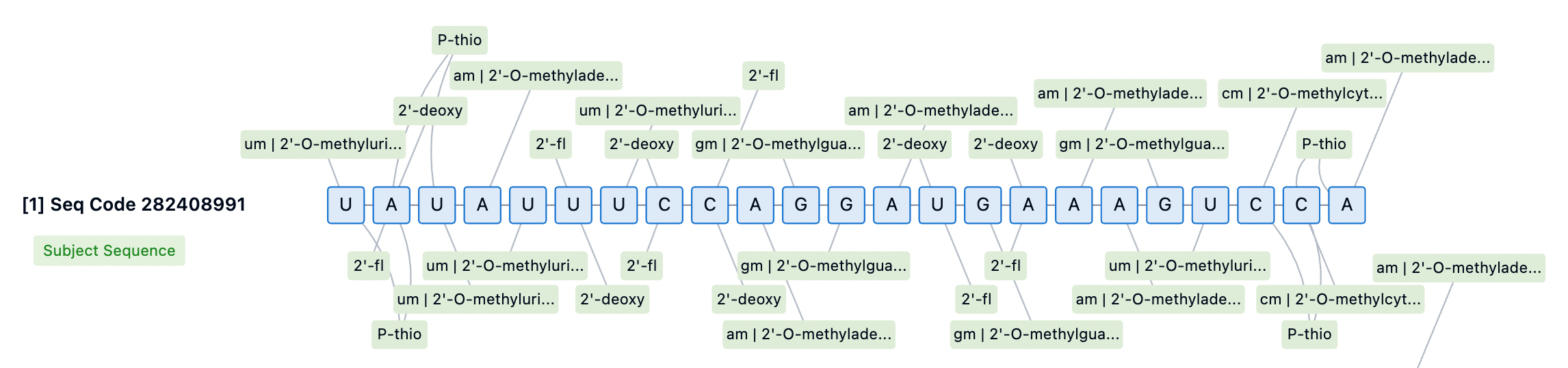
Stability of siRNA Drugs Against Nuclease Exonucleases
The long-term efficacy of siRNA drugs relies on the sustained release of the drug from endosomal and lysosomal compartments, as well as competition with naturally occurring microRNA (miRNA) for loading into newly synthesized Ago2. Even a slight decrease in RISC entry capability can negatively impact long-term biological efficacy. Therefore, extensive modifications to the siRNA scaffold are crucial for long-term stability, and the clinical durability differences among various approved drugs are at least partly due to variations in the 2'-OMe and 2'-F patterns used.
Nuclease exonuclease stability is a key consideration in siRNA drug design, as these enzymes can effectively degrade RNA and DNA independently of 2'-OH recognition. To enhance siRNA stability, scientists have employed a range of chemical modification strategies. Among these, 2'-O-methyl (2'-OMe) and 2'-fluoro (2'-F) modifications are effective methods for protecting siRNA from endonuclease-mediated cleavage. These modifications replace the 2'-OH group, enhancing siRNA's resistance to nucleases while maintaining the A-form helical structure of the guide strand, which is crucial for successful RISC recognition and loading. For example, through Patsnap Bio, we found that Givosiran incorporates extensive chemical modifications, including 2'-OMe and 2'-F modifications, as well as phosphorothioate (PS) modifications, all of which enhance siRNA stability, reduce immune responses, and improve cellular uptake and efficacy.
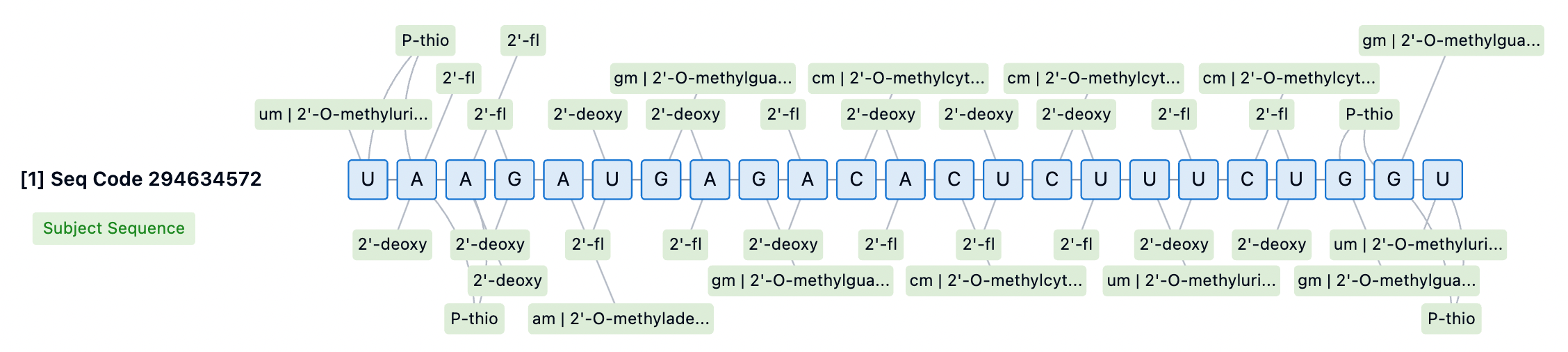
Additionally, phosphorothioate (PS) modifications are a primary strategy for improving the stability of siRNA at the 5' and 3' ends. Adding PS modifications to the backbone bonds at the 5' end can significantly enhance exonuclease stability, marking a key distinction between Alnylam's STC and ESC platforms. For instance, first-generation GalNAc-conjugated siRNAs, such as Revusiran, lacked these additional stability modifications, leading to cumulative toxicity issues in clinical trials. In contrast, newer-generation siRNA drugs like Inclisiran employ broader PS modifications to enhance siRNA stability and efficacy.
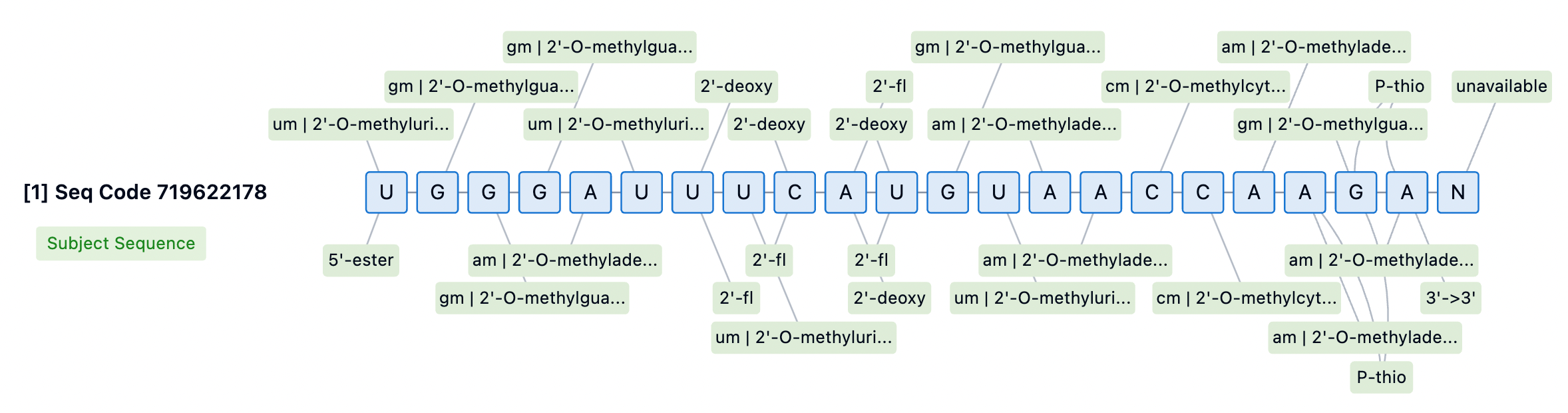
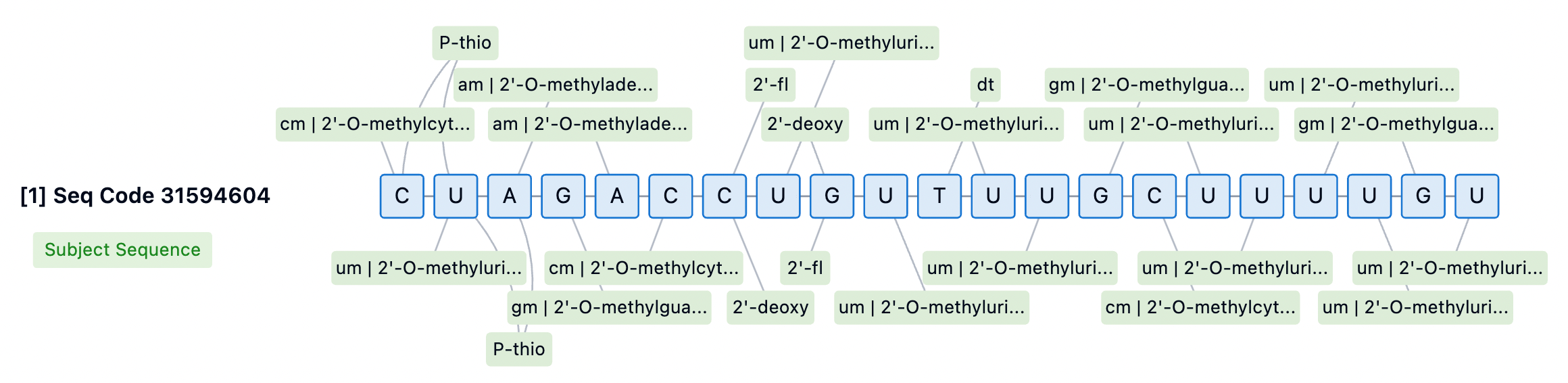
The 3' end of the siRNA passenger strand is typically conjugated to targeting moieties, providing some protection against 3' exonuclease-mediated degradation. However, none of the currently approved siRNA drugs incorporate PS modifications at the 3' end of the passenger strand. For internally or 5'-conjugated siRNA, stabilizing the 3' end is essential to prevent degradation by 3' exonucleases. Designs lacking PS at the 3' end may aid in releasing siRNA from membranes for RISC loading but could also diminish the durability of the siRNA. Thus, introducing cleavable linkers may be a strategy to improve conjugate release and 3' stability.
Summary
The development of siRNA drugs is a complex process that involves several key stages, including proof of concept (POC), clinical POC, and regulatory approval. In this process, chemical modifications play a crucial role in enhancing the stability of siRNA, reducing immunogenicity and off-target effects, and improving cellular uptake. These modifications include phosphorothioate modifications, ribose modifications, and base modifications, which can enhance the resistance of siRNA to nucleases, increase its affinity for target mRNA, and minimize immune responses and nonspecific binding.
Approved siRNA drugs, such as Alnylam's Onpattro (Patisiran), demonstrate the importance of chemical modifications in improving siRNA stability and efficacy. Patisiran incorporates 2'-O-methyl (2'-OMe) and 2'-fluoro (2'-F) modifications, as well as phosphorothioate (PS) modifications, which enhance siRNA stability, reduce immune responses, and improve cellular uptake and potency. Furthermore, the long-term efficacy of siRNA drugs relies on the sustained release of the drug from endosomal and lysosomal compartments, as well as competition with naturally occurring microRNA (miRNA) for loading into newly synthesized Ago2. Even a slight decrease in RISC entry capability can negatively impact long-term biological efficacy. Therefore, extensive modifications to the siRNA scaffold are essential for long-term stability, and the differences in clinical durability among various approved drugs are at least partly due to variations in the 2'-OMe and 2'-F patterns used.
Patsnap Bio, as a powerful biological sequence retrieval and analysis system, provides researchers with a comprehensive resource to support the discovery and development of siRNA drugs. By manually annotating key biological sequences and marking modification structures, the efficiency of obtaining sequence information is enhanced. Additionally, the knowledge graph associates biological sequences with multidimensional information about species, drugs, diseases, structures, and functions, which is crucial for non-clinical pharmacodynamics research and proof of concept studies for siRNA drugs, aiding in understanding their mechanisms of action and guiding clinical trial design.
Better answers for better bio-innovations!
Validate novelty, eliminate risk, and innovate with confidence using the world’s largest sequence database curated from millions of patent and non-patent sources.
Patsnap Bio helps you turn weeks into minutes with cutting-edge AI-enabled tools built to master the complexities of sequence retrieval and automate IP analysis with precision and ease.
With best-in-class coverage of protein and nucleic acid sequences combined with state-of- the-art search algorithms, you’ll spend less time searching and more time bringing your bio-innovations to market.
Reference
1.Hu, B. et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther 5, 101 (2020). https://s10-doi-org.libproxy1.nus.edu.sg/s41392-020-0207-x
2.Tang, Q. & Khvorova, A. RNAi-based drug design: considerations and future directions. Nat Rev Drug Discov 23, 341-364 (2024). https://s10-doi-org.libproxy1.nus.edu.sg/s41573-024-00912-9




