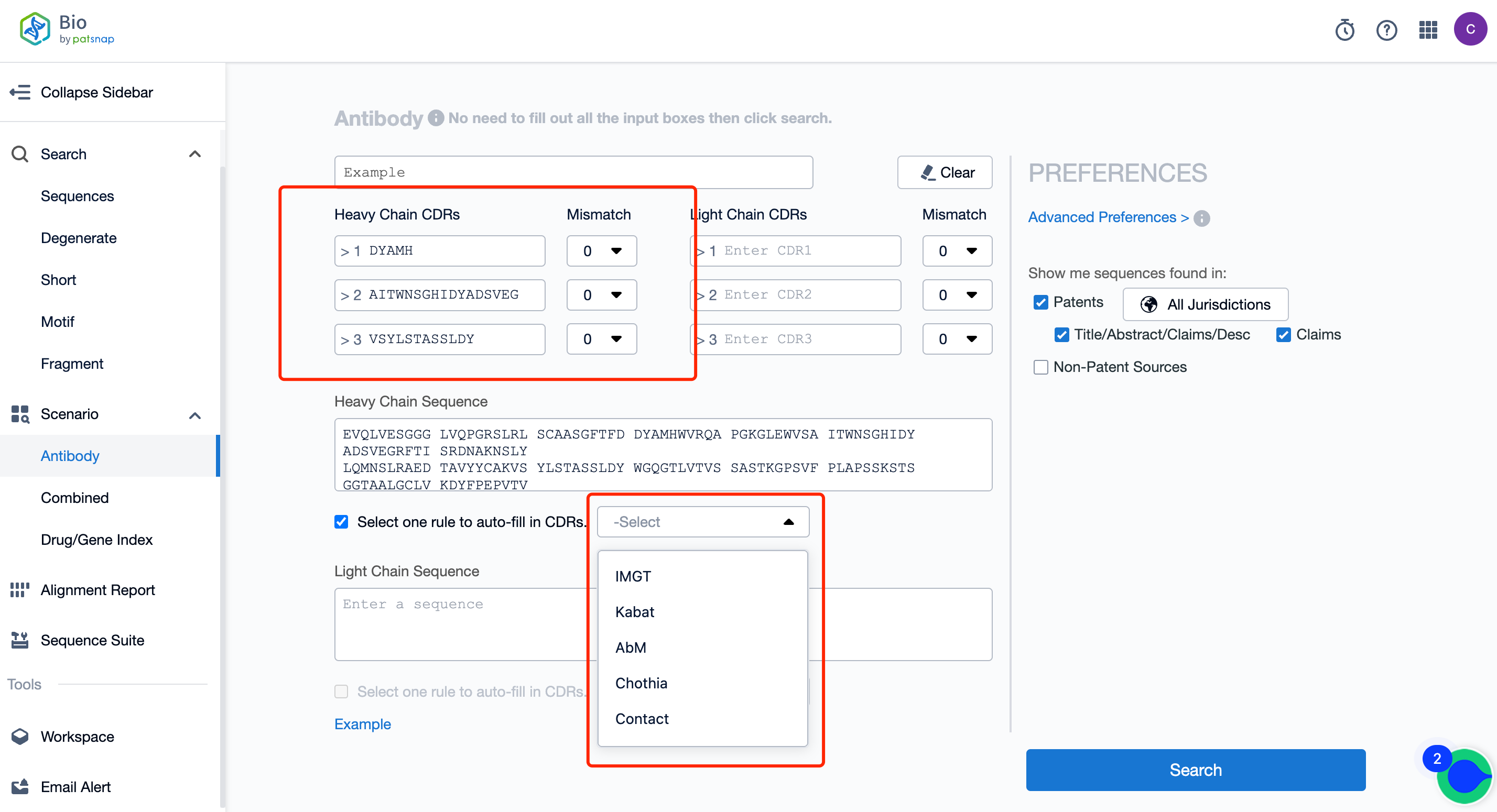
Is it sufficient to use only one encoding rule for the retrieval of antibody CDR sequences in FTO?
To reduce the risk of missing FTO sequences in antibody searches, it is insufficient to use only one encoding rule for CDR sequences. Antibody CDR sequences have multiple encoding rules, including the common ones such as Kabat, IMGT, Chothia, etc., and the CDR sequences under different encoding rules are not consistent. Therefore, to ensure the comprehensiveness of the FTO search and reduce the risk of omissions, at least two or more encoding rules should be selected. It is commonly suggested in this field that the CDR sequences under the Kabat, IMGT, and Chothia codes should be searched.
Here, it is recommended to use the Patsnap Bio Sequence Database for antibody sequence retrieval. This database supports the input of CDR sequences under five coding rules, which is convenient and greatly improves the work efficiency of the searchers.
It is important to note that Patsnap Bio is the most extensive sequence search platform for the Patsnap database. It incorporates AI with human-curated data for comprehensive handling of protein and nucleotide sequence data plucked from global patents, biological periodicals, and public repositories. Essential biological sequences are manually annotated, illuminating structural modifications to provide the most accurate sequence data and boost sequence retrieval efficiency.
Free registration is available for the Bio biological sequence database: https://bio-patsnap-com.libproxy1.nus.edu.sg. Act now to expedite your sequence search tasks.