A Retrieval Scheme for Any Combination of Antibodies Light Chains, Heavy Chains, and CDR
Patsnap Bio Sequence Database can retrieve antibody light and heavy chains, CDR sequence, and any CDR combination.
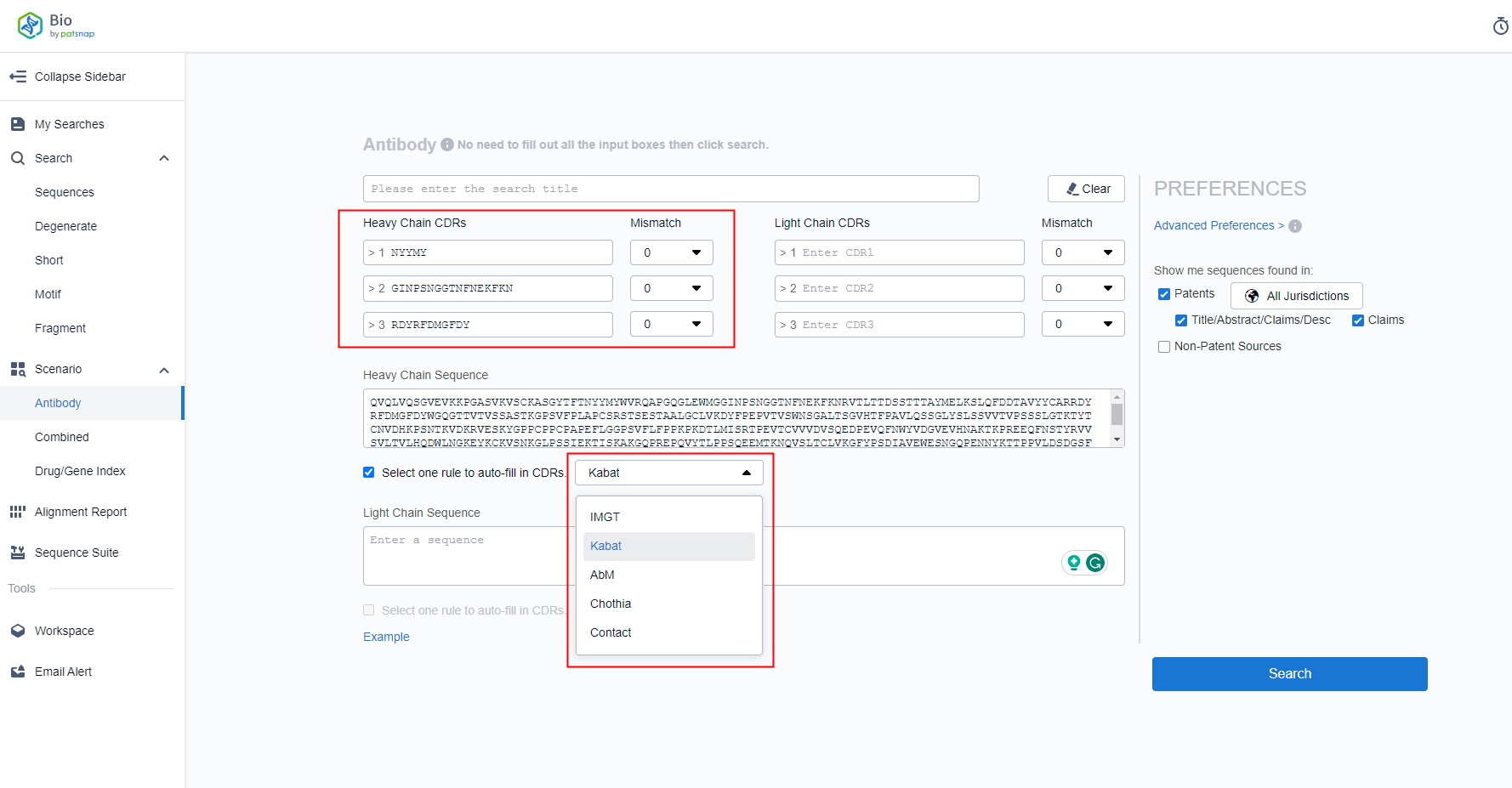
On the one hand, one or more light and heavy chain CDR sequences can be retrieved by manual input. On the other hand, by inputting the full-length sequence of light and heavy chains and clicking on the encoding rules, the corresponding CDR sequence can be automatically filled in (see Figure 1).
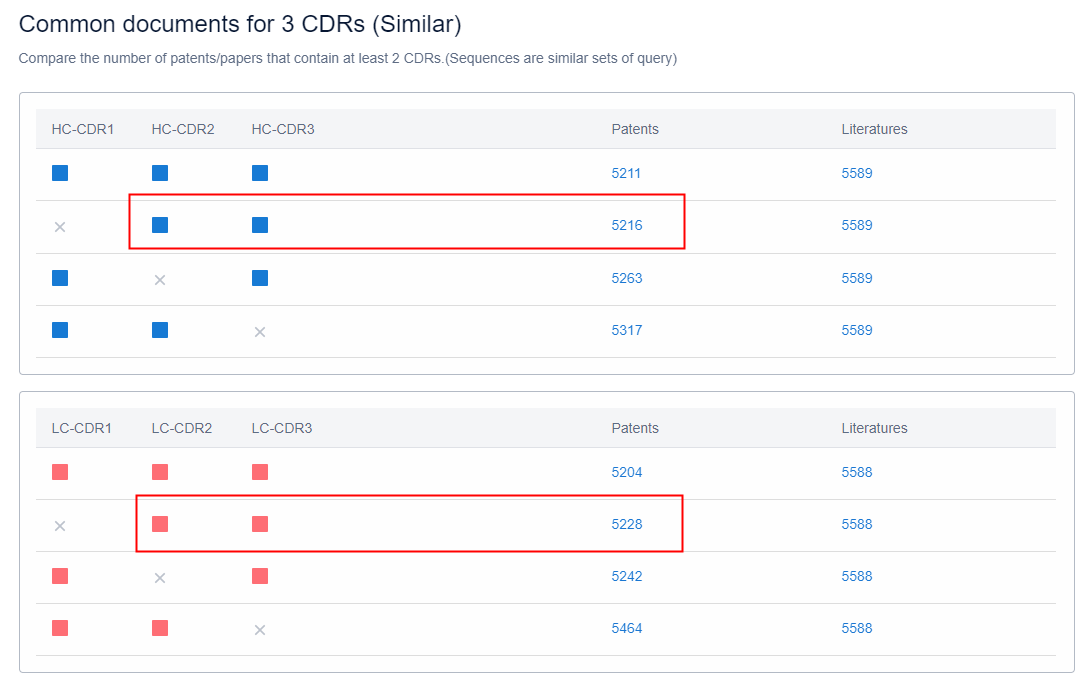
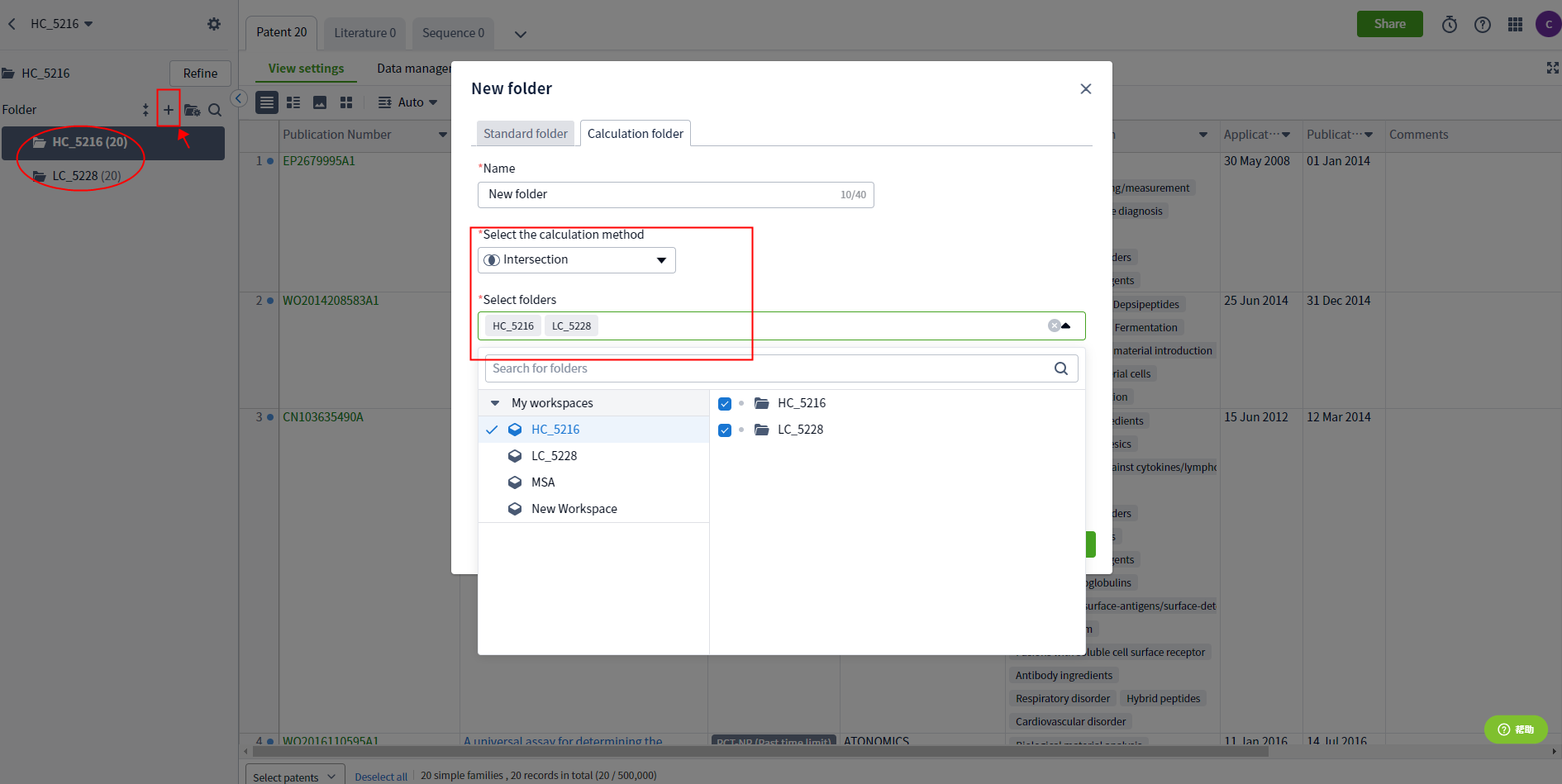
Moreover, on the "View Sources" page of the sequence retrieval results, you can see the public source results of any CDR sequence combination, and you can view the corresponding patents/documents by clicking on the results. If you want to see the common public patents of HCDR2, HCDR3, LCDR2, and LCDR3 (see Figure 2), save patents 5216 and 5228 to two folders in the workspace, and then take the intersection of the two folders to get the common public results (see Figure 3). The same method can be used to find common public patents of other combinations.
It is important to note that Patsnap Bio is the most extensive sequence search platform for the Patsnap database. It incorporates AI with human-curated data for comprehensive handling of protein and nucleotide sequence data plucked from global patents, biological periodicals, and public repositories. Essential biological sequences are manually annotated illuminating structural modifications to provide the most accurate sequence data and boost sequence retrieval efficiency.
Free registration is currently available to utilize the Bio biological sequence database: https://bio-patsnap-com.libproxy1.nus.edu.sg. Act now to expedite your sequence search tasks.