Reducing the Risk of FTO Missed Detection in Special Referential Sequences of Patents
The patent and literature contain sequences of nucleic acids containing N and protein sequences containing X. I'm interested in reducing the risk of missing such sequences. Is there a recommended database where I can search for these sequences?
These generic sequences found in patents contain a large number of degenerate symbols. Traditional search algorithms need help to identify them due to their low similarity scores. Here, I recommend the Patsnap Bio Sequence Database. This database features an exclusive degenerate search algorithm.
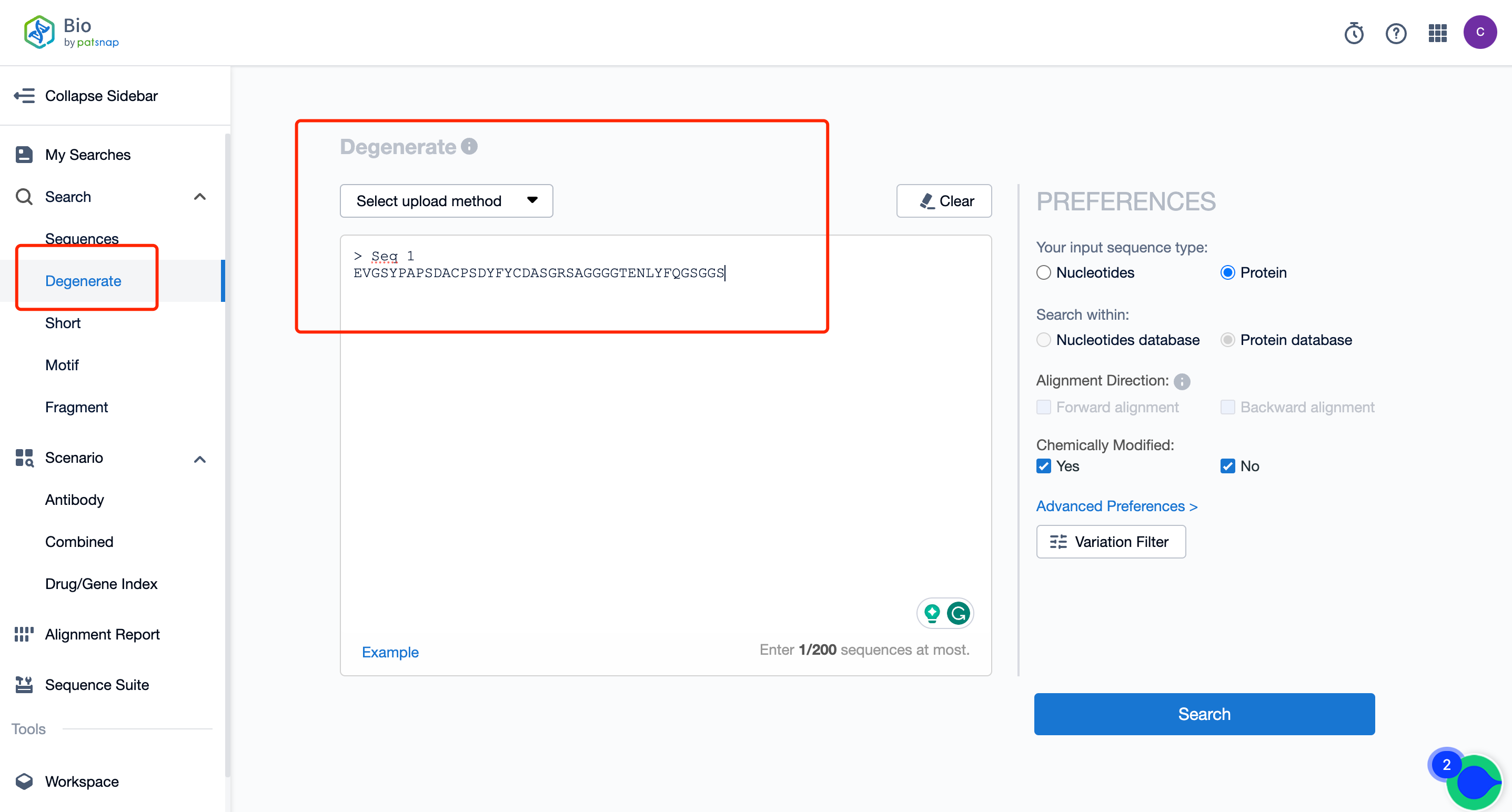
To begin with, click here to register for a free account on the Bio database. Once you're on the database homepage, click the “Degenerate” option (Figure 1) and enter the sequence you're interested in.
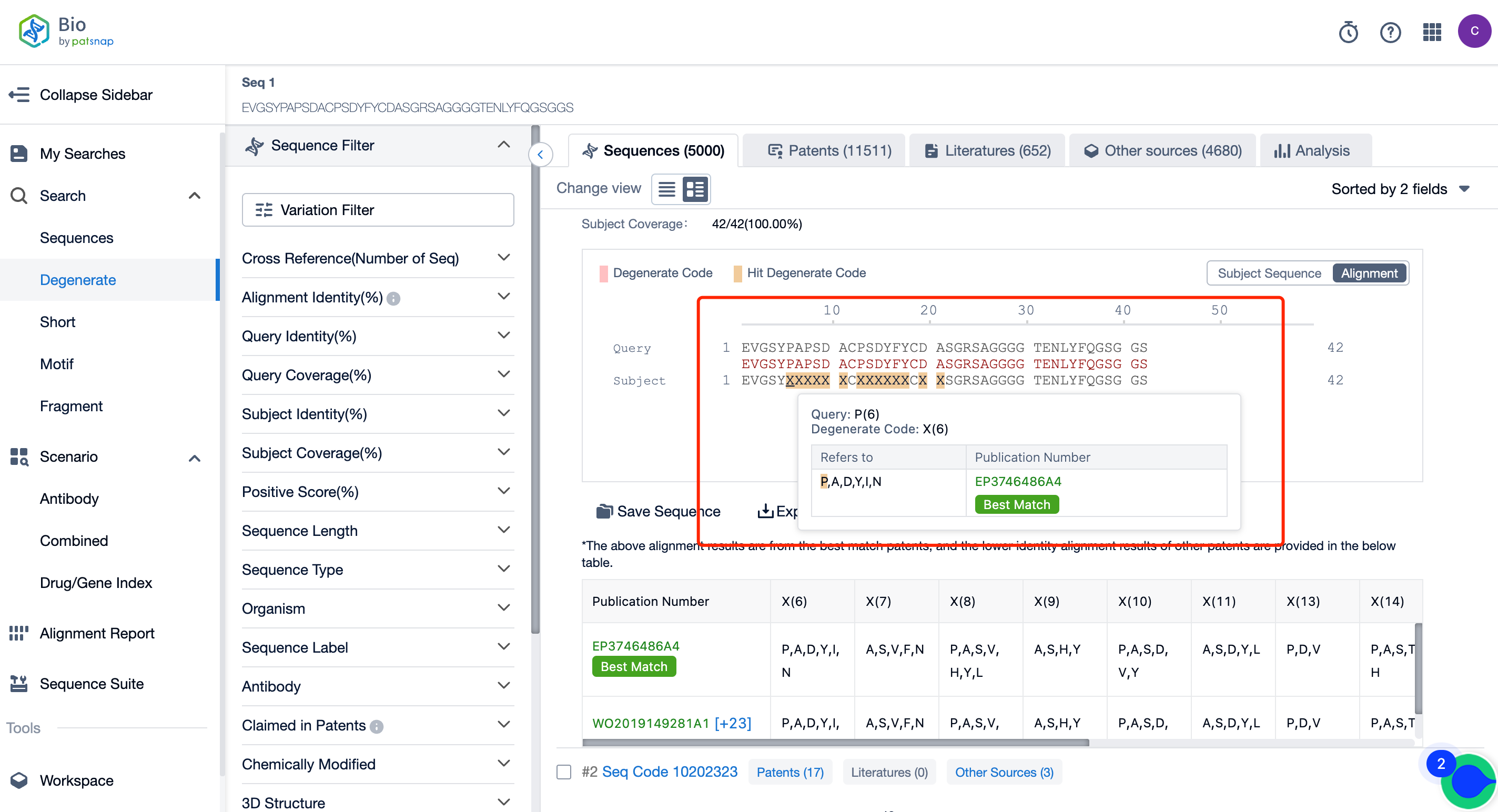
After initiating the search, you'll arrive at the search results page (Figure 2). Here, you can compare the sequences that match with ones containing X in the generic format. By hovering your mouse over X, you can also access information about its representation in the patent documentation, which is extremely helpful. This generic search algorithm significantly reduces the risk of overlooking these sequences, making it highly recommended for your needs.
It is important to note that Patsnap Bio is the most extensive sequence search platform for the Patsnap database. It incorporates AI with human-curated data for comprehensive handling of protein and nucleotide sequence data plucked from global patents, biological periodicals, and public repositories. Essential biological sequences are manually annotated, illuminating structural modifications to provide the most accurate sequence data and boost sequence retrieval efficiency.
Free registration is available for the Bio biological sequence database: https://bio-patsnap-com.libproxy1.nus.edu.sg. Act now to expedite your sequence search tasks.