Step-by-Step Guide on How to Search for Homologous Sequences of a Specific Gene
Homologous sequences refer to two sequences that share a common ancestor. Homology is a qualitative concept where we determine whether two sequences are homologous or not. However, the similarity between homologous sequences can be measured using techniques such as sequence alignment. One popular method for aligning sequences is using BLAST (Basic Local Alignment Search Tool).
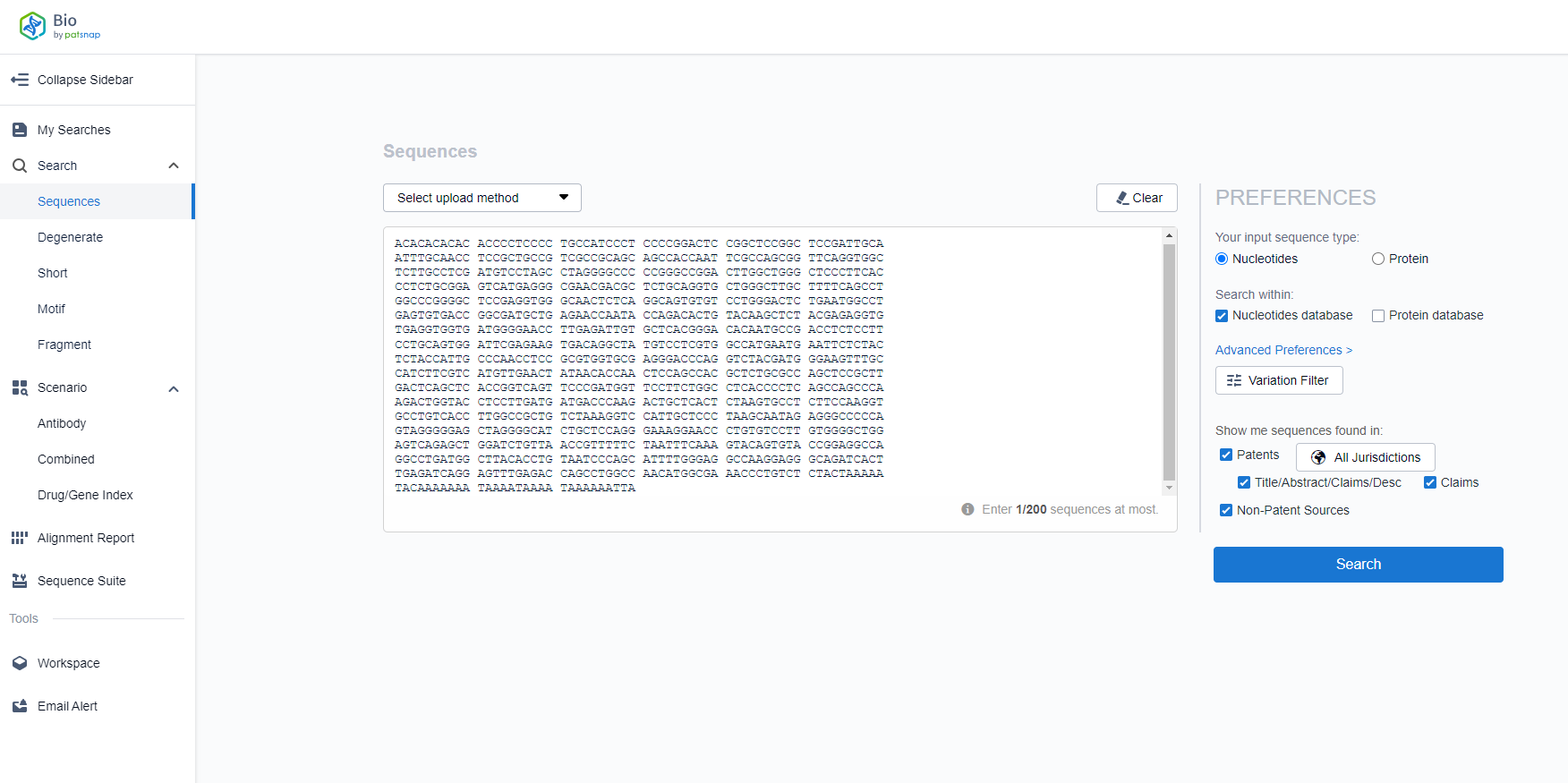
The Patsnap Bio Sequence Database includes sequences from patents, literature, and third-party sources, making it the most comprehensive sequence database in the world. By performing sequence alignment, one can quickly and accurately identify homologous sequences of a gene. Taking the example of the human ERBB3 gene sequence, you can search for it in the Patsnap Bio sequence database as shown in the figure below:
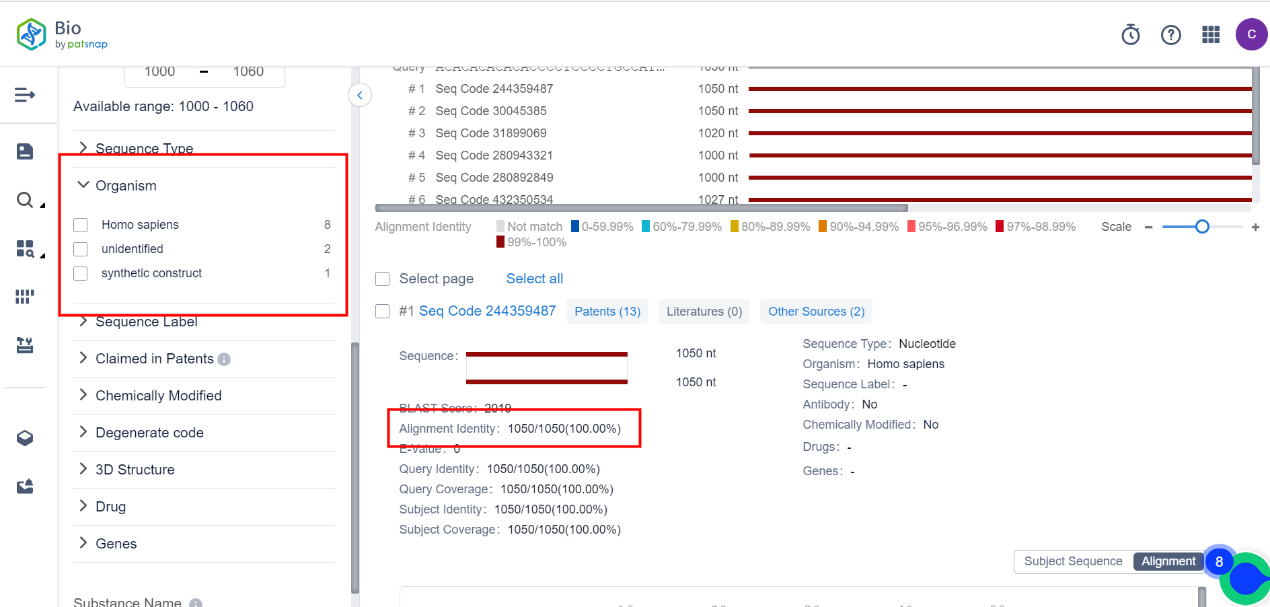
By examining the alignment results of the gene sequence, if you want to see whether the gene sequences from specific organisms have homology with the query sequence, you can use the species filter on the left side to perform filtering, as shown in the figure below. You can also sort the results by similarity to quickly and accurately find highly similar homologous sequences.
It is important to note that Patsnap Bio is the most extensive sequence search platform for the Patsnap database. It incorporates AI with human-curated data for comprehensive handling of protein and nucleotide sequence data plucked from global patents, biological periodicals, and public repositories. Essential biological sequences are manually annotated illuminating structural modifications to provide the most accurate sequence data and boost sequence retrieval efficiency.
Free registration is currently available to utilize the Bio biological sequence database: https://bio-patsnap-com.libproxy1.nus.edu.sg. Act now to expedite your sequence search tasks.